Projects
PubMed Antibiotic Resistance Trend Analysis Dashboard
Project Overview
This project presents an interactive web dashboard developed with Streamlit, designed to analyze and visualize trends in scientific publications related to Antibiotic Resistance (AR) sourced from PubMed. Leveraging Natural Language Processing (NLP) and Topic Modeling (LDA), this tool provides key insights into research evolution, prevalent themes, and emerging areas within the field of antibiotic resistance.
Available at: https://amrtrendanalysis-zoajgryzuojpr3f9atwicc.streamlit.app/
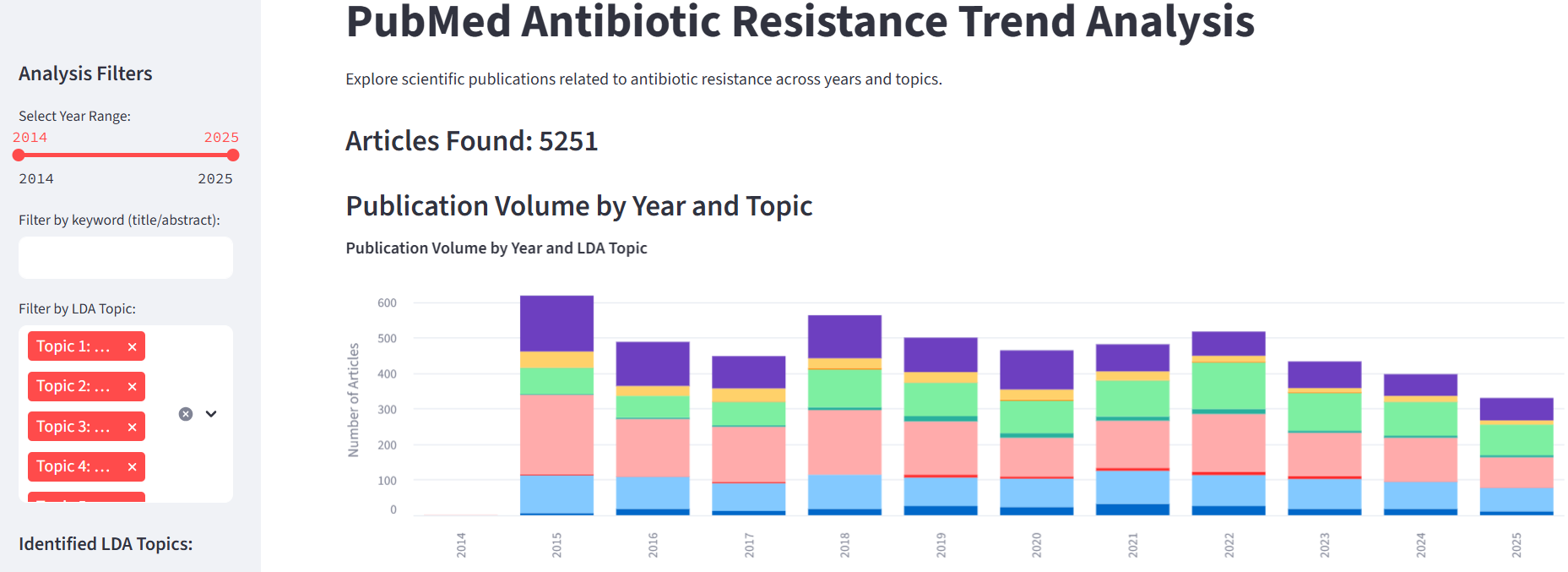
This dashboard demonstrates my ability to:
- Acquire and process real-world, unstructured data.
- Apply advanced NLP techniques (including text cleaning, tokenization, and Latent Dirichlet Allocation for topic modeling).
- Extract meaningful insights from large text datasets.
- Develop interactive data visualizations for clear communication of complex findings.
- Build deployable data applications.
Key Features
- Interactive Filtering: Filter publications by publication year range and keywords within titles/abstracts.
- Dynamic LDA Topic Analysis: Explore the dominant research themes automatically identified from the text data. Each topic is accompanied by a descriptive label derived from its most prominent keywords.
- Temporal Trend Visualization: Observe the evolution of publication volume over time, broken down by identified LDA topics, showcasing shifts in research focus.
- Topic Distribution Overview: Understand the overall prevalence of each research topic within the filtered dataset.
- Word Cloud Generation: Visualize the most frequent keywords for the currently filtered articles, offering a quick grasp of key terminology.
Technologies Used
- Python: Core programming language.
- Pandas: Data manipulation and analysis.
- Streamlit: For building the interactive web dashboard.
- Altair: Declarative statistical visualizations.
- NLTK / SpaCy (Implicit via NLP Pipeline): Text preprocessing (tokenization, stopwords, lemmatization).
- Gensim (Implicit via NLP Pipeline): Implementation of Latent Dirichlet Allocation (LDA) for topic modeling.
- WordCloud: Generating visual word clouds.
- Matplotlib: For displaying Word Clouds.
Word Cloud
A picture is worth a thousand words. One interesting way to summarize the content of a text is through word clouds. In this type of visualization, the most frequent words in a text are displayed in larger font size.
In this script, we summarize the content of the current Chilean Constitution, as well as the proposed 2023 Constitution.
Here is the word cloud of the current document:

And the word cloud for the 2023 proposal:
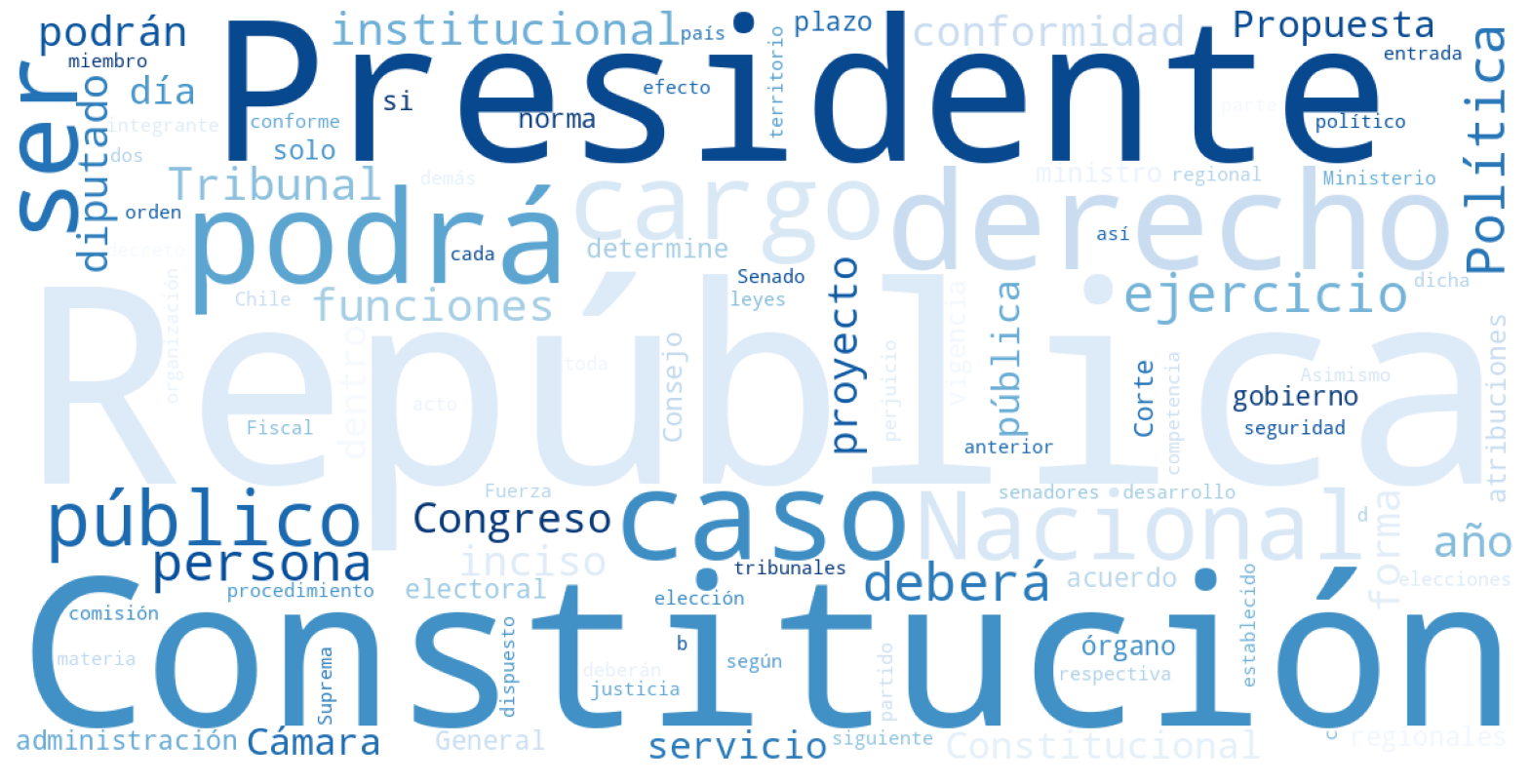 What changes can you notice?
If you want to learn how to generate your own word cloud from any PDF file, check out the script. here
What changes can you notice?
If you want to learn how to generate your own word cloud from any PDF file, check out the script. here
Impact of Transfer Learning on Image Classification with Artificial Intelligence
Artificial Intelligence (AI) has shown its potential in multiple fields, including image classification. Massive models such as ResNet50 can classify thousands of distinct regular entities, such as cars or animals, and were trained on millions of labeled images. However, their applications can go beyond their initial goals through a methodology known as Transfer Learning (TL). With TL, we can redirect a powerful model like ResNet50 toward a classification problem of our own interest.
Below, we use this model implemented in the Keras library, applying TL to classify two types of data.
First, we classify histological images of normal and cancerous colon tissue.
Then, we apply this method to classify images of high- and low-quality lemons.
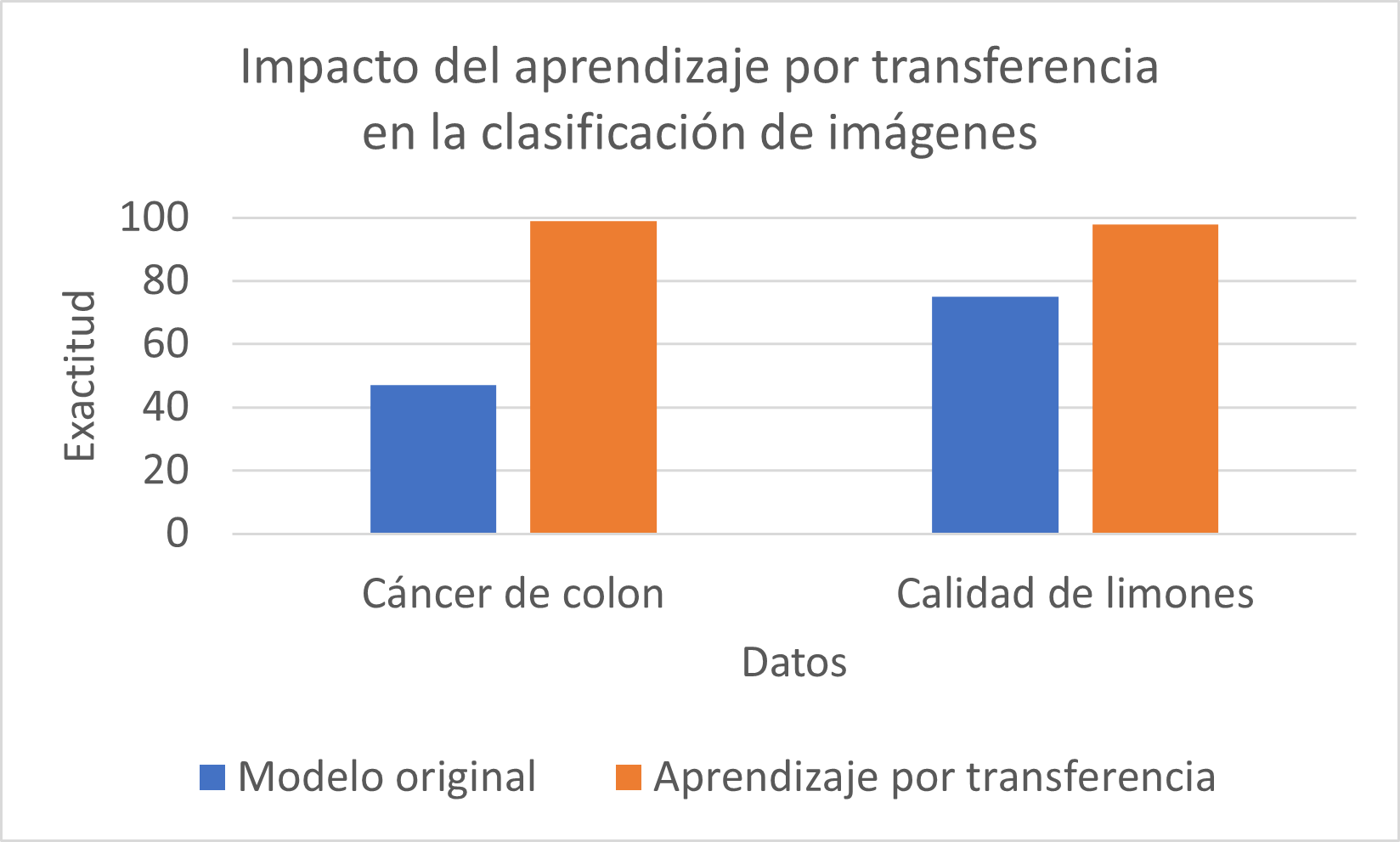
In the chart, we can see how applying transfer learning allows the ResNet50 model to improve from 47% to 99% accuracy when classifying histological images of normal and cancerous colon tissue.
Similarly, by applying transfer learning, the model improves from 75% to 98% accuracy when classifying images of high- and low-quality lemons.
The potential of this type of methodology is undeniably impressive.
Identity Matrix
Determining sequence identity is key. It helps us understand the evolutionary relationships between isolates or infer the function of a new sequence, among other applications.
In the following Python script, we demonstrate how to apply local sequence alignments using the Smith-Waterman algorithm.
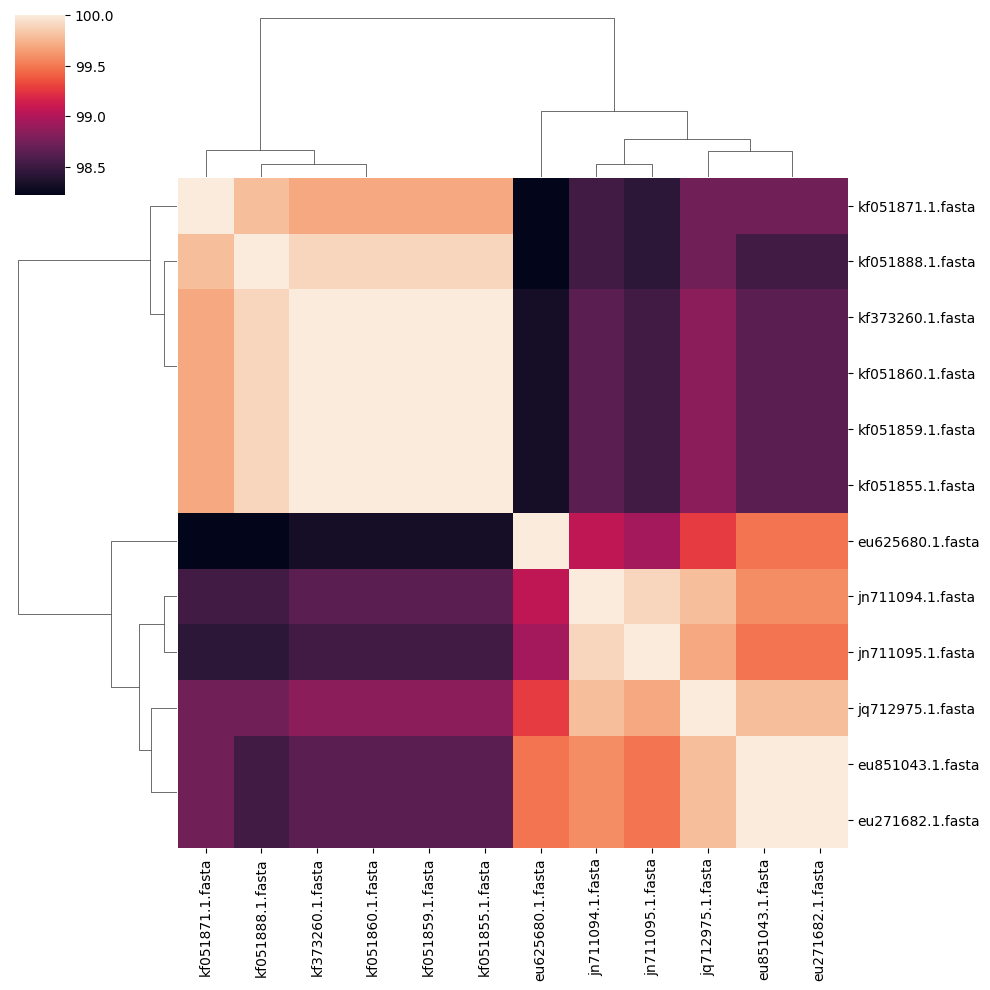
In the clustermap, two groups or clusters of sequences with higher identity stand out.
Which sequences are more identical to each other?
You can check which viruses they correspond to by copying their names (excluding the “.fasta” extension) and searching for them in the NCBI database.
Differential Expression
RNA sequencing using high-throughput technologies allows us to uncover expression patterns.
In the following R script, we demonstrate how to statistically analyze expression differences.
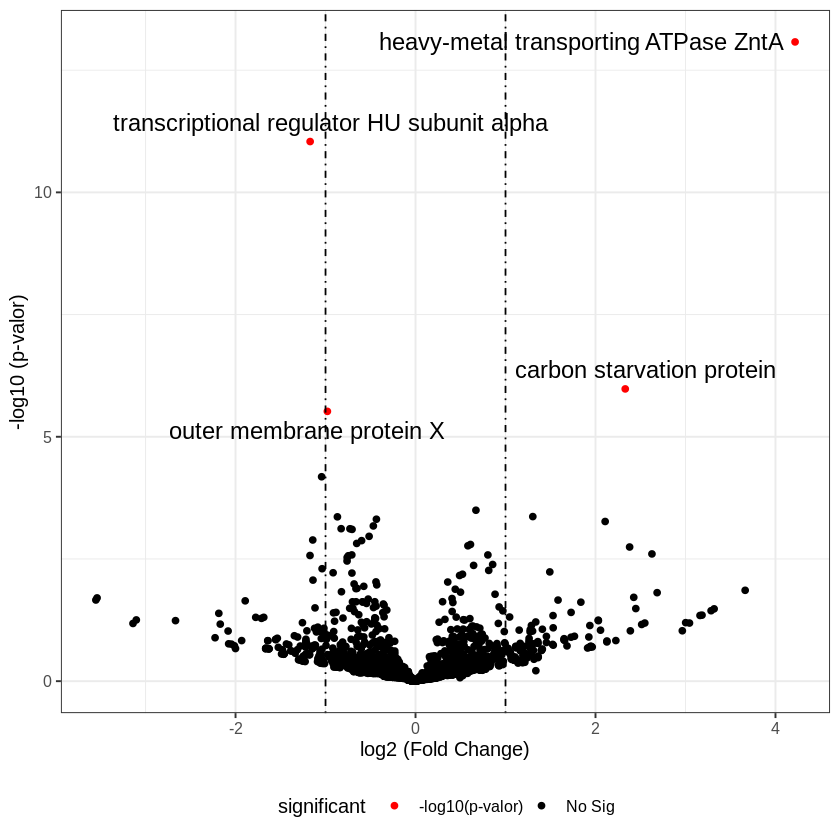
The results can be summarized in a volcano plot, where expression differences are represented as a fold change factor.
In the plot, points to the right of zero indicate genes with higher expression in the treatment condition compared to the control condition.
Points to the left of zero indicate the opposite, lower expression in the treatment condition compared to the control condition.
However, only the red points are considered statistically significant.
Which genes show differential expression according to the plot?
For more details about the data source, you can review the script.